A Python implementation of the Icoshift algorithm, a versatile tool for the rapid alignment of 1D NMR spectra
Icoshift is a Matlab-based algorithm for the alignment of NMR spectra developed by Francesco Savorani and Giorgio Tomasi. It performs correlation shifting of spectral intervals using an FFT engine that aligns all spectra simultaneously. I've personally found it incredibly useful in the processing of data, particularly through Metabolab.
The Matlab algorithm is demonstrated to be faster than similar methods found in the literature making full-resolution alignment of large datasets feasible and thus avoiding down-sampling steps such as binning. The algorithm uses missing values as a filling alternative in order to avoid spectral artifacts at the segment boundaries.
While extending the NMR spectra processing capabilities of MetaPath it became obvious that spectral alignment would be essential in the toolkit - and Icoshift was the obvious choice. While a Matlab bridge was in process, I wanted to see if it was possible to re-code the Icoshift algorith natively in Python - allowing MetaPath users to get access to it without having Matlab installed.
The algorithm was converted to Python using SMOP followed by hand re-coding using test datasets to check output at various steps. Better (and more complicated) test cases to come. The interface remains identical to the Matlab version at present.
Getting started
To install (assuming you have installed pip):
pip install icoshift
To use from your own script:
import icoshift
xCS,ints,ind,target=icoshift.icoshift('average',test)
Where test is an numpy.array of data - subjects in rows, ppm in columns. The outputs match those in the Matlab script: of most interest is xCS (the shifted spectra).
Here Be Dragons
Conversion from one programming language to another is not straightforward. Particularly problematic from MATLAB to Python is the change from zero-based to one-based indexing. The implementation has been fixed to work and produce comparable output for all inputs, however issues with some datasets or settings may remain. Full tests to confirm equivalence to the MATLAB algorithm to follow.
To support developers in [[ countryRegion ]] I give a [[ localizedDiscount[couponCode] ]]% discount on all books and courses.
[[ activeDiscount.description ]] I'm giving a [[ activeDiscount.discount ]]% discount on all books and courses.
But it works
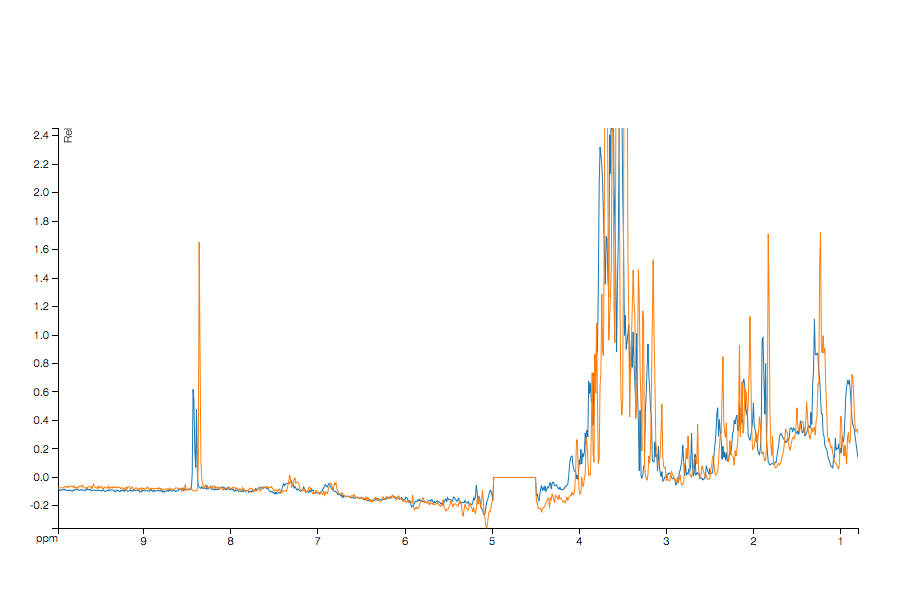
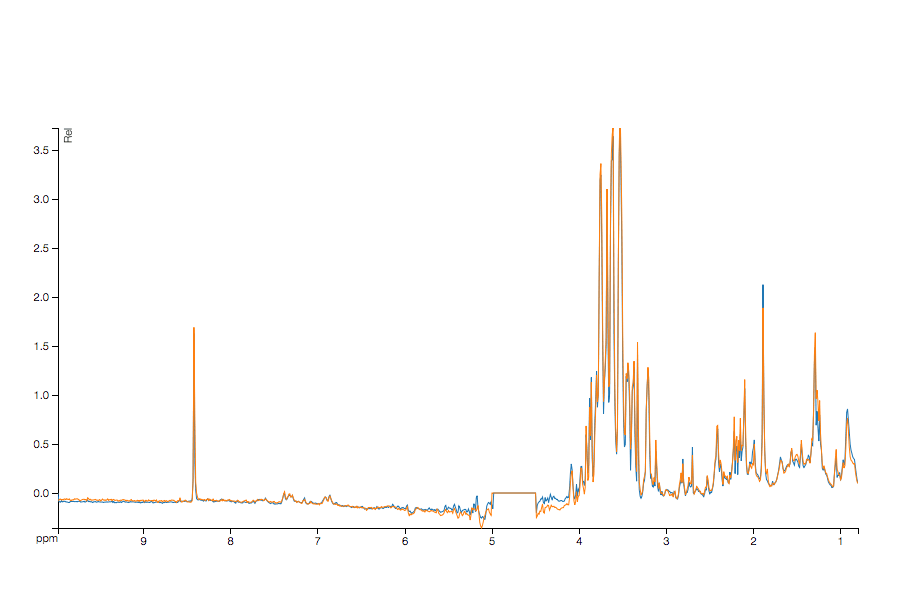
Here is some sample output (run through MetaPath - yes there is already a plugin) showing the original and shifted data from a sample manually off-shifted dataset.